Because this piece does not have an abstract, we have provided for your benefit the first 3 sentences of the full text.
To the Editor: Strong association of the extended major histocompatibility complex (xMHC) region on human chromosome 6 with schizophrenia has been supported by a number of genome-wide association studies. However, since the xMHC region is featured by numerous polymorphisms, dense gene clusters, and strong linkage disequilibrium, it is difficult to attribute the association to specific genes. A targeted scrutinization of gene expression in the xMHC region can provide valuable candidate genes for future validation.
Gene Expression Profiling of the xMHC Region Reveals 9 Candidate Genes in Schizophrenia
To the Editor: Strong association of the extended major histocompatibility complex (xMHC) region on human chromosome 6 with schizophrenia has been supported by a number of genome-wide association studies.1 However, since the xMHC region is featured by numerous polymorphisms, dense gene clusters, and strong linkage disequilibrium, it is difficult to attribute the association to specific genes. A targeted scrutinization of gene expression in the xMHC region can provide valuable candidate genes for future validation. Here we utilize 2 real-time polymerase chain reaction (PCR)-based platforms and 2 sample sets to investigate protein-coding gene expressions in the xMHC region in peripheral blood leukocytes to identify schizophrenia candidate genes.
Methods. The 2 sample sets include 1 discovery sample set (24 patients and 24 healthy controls) and 1 validation sample set (41 patients and 37 controls) (Supplementary eTable 1). All patients were first-episode, drug-naׯve, and diagnosed with schizophrenia according to the Fourth Edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV). All patients and controls were recruited after informed consent and institutional review board approval. Blood samples were collected in the morning after overnight fasting (10-12 hours). Leukocytes were isolated, and RNA was extracted and reverse transcribed into complementary DNA. First, we applied real-time PCR-based TaqMan low-density arrays (TLDAs) (Thermo Fisher Scientific) to screen differential gene expressions in the discovery sample set. Expressions of 162 protein-coding genes located in the xMHC region were investigated, with expression of ACTB (actin, beta) as an internal standard. Second, we reprepared RNA samples in the discovery set and reassayed previous positive genes by 384-well plate TaqMan real-time PCR. Third, we evaluated the result in the independent validation sample set by 384-well plate TaqMan real-time PCR.
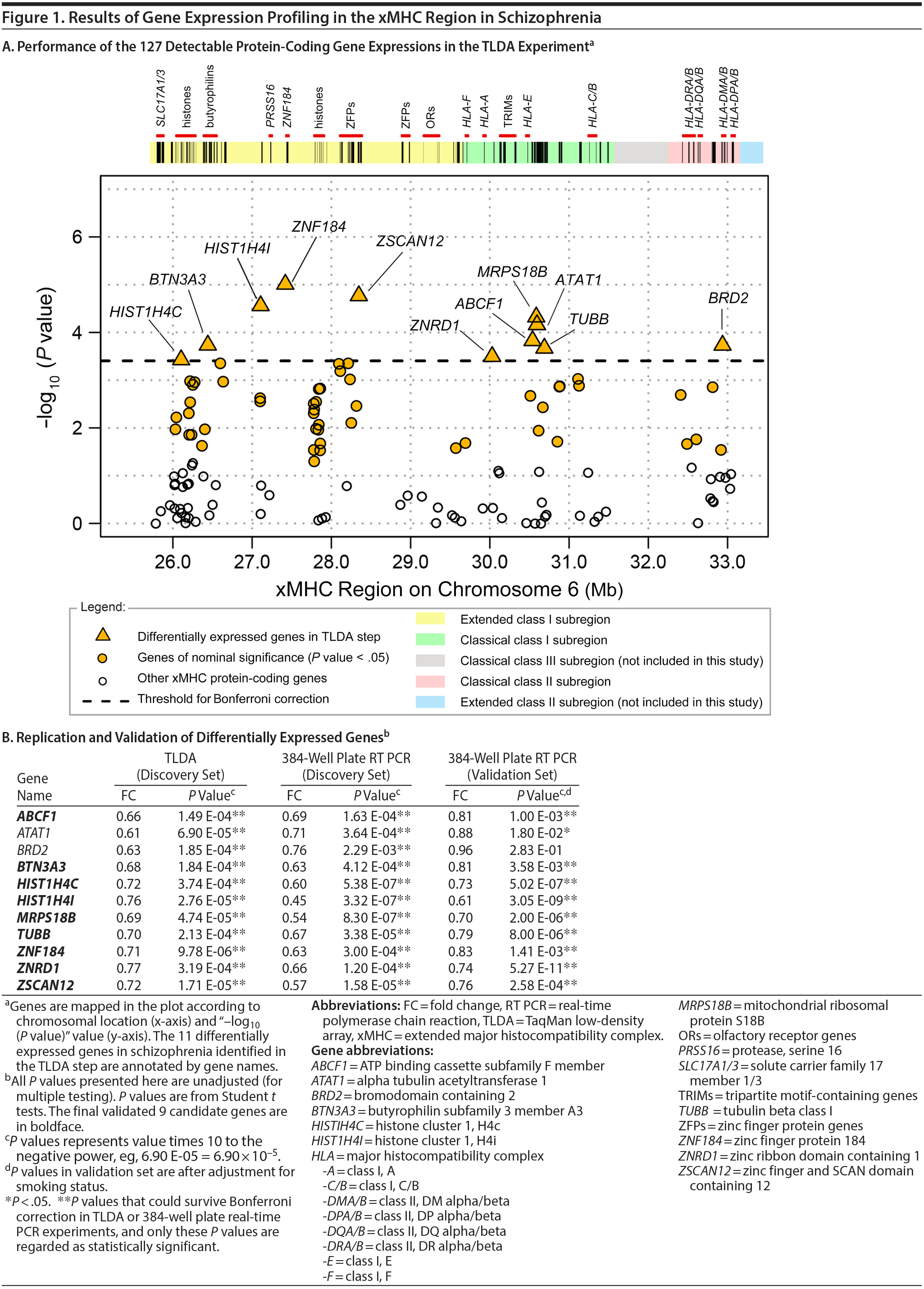
Results. In the TLDA experiment, 127 of the 162 protein-coding genes were above the limit of detection. Sixty genes reached nominal significance in Student t tests (unadjusted P values < .05), while 11 of them survived Bonferroni correction for multiple comparisons and thus were regarded as differentially expressed genes in this step (Figure 1A). As shown in Figure 1B, expression of the 11 genes were all significantly decreased in schizophrenia with prominent fold changes (P values: 9.78 ×— 10−6 to 3.74 ×— 10−4, fold changes: 0.61 to 0.77). This result was successfully confirmed with the 384-well plate TaqMan real-time PCR method, which supported even greater changes (P values: 3.32 ×— 10−7 to 2.29 ×— 10−3, fold changes: 0.45 to 0.76). In the independent validation sample set, the 11 genes consistently exhibited decreased trend of expressions in schizophrenia, among which 9 genes with statistical significance were finally identified as candidate genes (P values: 5.27 ×— 10−11 to 3.58 ×— 10−3 [after adjustment for smoking status], fold changes: 0.61 to 0.83).
In the 9 validated genes,* there were 3 genes that encode zinc-binding proteins, ie, ZNF184, ZSCAN12, and ZNRD1. Zinc-binding proteins usually regulate transcriptions. It deserves future investigation to determine whether and how these genes would affect gene expressions in schizophrenia. Genes HIST1H4I and HIST1H4C encode histones. Strong evidence of associations was observed within and around histone genes in schizophrenia.2 Abnormal expression of genes related to nucleosome and histone structure and function has also been found in both schizophrenia patients and their siblings.3 MRPS18B encodes ribosomal protein that helps in mitochondrial protein synthesis. TUBB encodes tubulin that is the major constituent of microtubules. Altered expression of TUBB has been reported in a previous microarray study in schizophrenia patients.4 ABCF1 and BTN3A3 are immune-related genes. BTN3A3 is involved in T-cell activity in adaptive immune response. ABCF1 enhances protein synthesis and promotes inflammation. Decreased expression of ABCF1 had been found in the whole blood of patients with schizophrenia and identified as a hub gene in a gene expressional subnetwork.5 Decreased expression of ABCF1 and BTN3A3 genes implies malfunctioning of inflammation processes in schizophrenia. Although changes in peripheral blood have limited power to directly reflect changes in brain tissues due to blood-brain barrier or tissue specificity, disturbances in the main neurotransmitter and hormonal systems in the central nervous system are concomitant with altered function of blood immune cells.6 Moreover, a recent study7 reported the discovery of the lymphatic vessel system in the central nervous system, which suggests gene expression changes in blood leukocytes, might affect normal inflammation process in brains and cause psychiatric symptoms.
In conclusion, this study revealed down-regulation of 9 xMHC-region gene expressions in first-episode and drug-naׯve schizophrenia patients, which was consolidated by 2 real-time PCR-based experimental platforms and 2 independent sample sets. This result suggests a decreased transcriptional activation in the xMHC region and highlights potential malfunctioning of immune cells in schizophrenia. Compounds that can normalize the expressions of certain genes may have therapeutic effects. Although our sample sizes are relatively small, given the strictly controlled sample background and the consistency in 2 subsets, the candidate genes we identified may serve as genetic markers for schizophrenia. Future validation in other larger populations is suggested.
*Definitions for the genes appear in the Figure 1 footnote.
References
1. Corvin A, Morris DW. Genome-wide association studies: findings at the major histocompatibility complex locus in psychosis. Biol Psychiatry. 2014;75(4):276-283. PubMed
2. Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753-757. PubMed
3. Glatt SJ, Stone WS, Nossova N, et al. Similarities and differences in peripheral blood gene-expression signatures of individuals with schizophrenia and their first-degree biological relatives. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(8):869-887. PubMed doi:10.1002/ajmg.b.31239
4. de Jong S, van Eijk KR, Zeegers DW, et al. PGC Schizophrenia (GWAS) Consortium. Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur J Hum Genet. 2012;20(9):1004-1008. PubMed doi:10.1038/ejhg.2012.38
5. de Jong S, Boks MP, Fuller TF, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS ONE. 2012;7(6):e39498. PubMed doi:10.1371/journal.pone.0039498
6. Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28(3):559-576. PubMed doi:10.1016/j.pnpbp.2004.01.009
7. Louveau A, Smirnov I, Keyes TJ, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337-341. PubMed
aBio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Key Laboratory of Translational Psychiatry, Shanghai Mental Health Center, Shanghai Jiao Tong University, Shanghai, China
bInnovation Center China, Asia & Emerging Market iMed, AstraZeneca Innovation Medicines and Early Development, Shanghai, China
cMental Health Institute of the Second Xiangya Hospital, Central South University, Key Laboratory of Psychiatry and Mental Health of Hunan Province, Hunan Province Technology Institute of Psychiatry, Changsha, China
‘ ¡Dr Sun and Ms Qing contributed equally to this letter.
*Dr Tan and Dr Wan contributed equally to this letter, and both qualify to serve as corresponding authors.
Potential conflicts of interest: None.
Funding/support: This work was supported by the 973 Program (2012CB910100) and the National Nature Science Foundation of China (81271486).
Role of the sponsor: The funding organizations are public institutions and had no role in the design and conduct of the study; collection, management, and analysis of the data; or preparation, review, and approval of the manuscript.
Acknowledgment: The authors thank Jinghong Chen, MM, and Xuemei Peng, MM, for their contributions in participant recruiting and sample pre-processing in this study. Both were from Mental Health Institute of the Second Xiangya Hospital, Central South University, Changsha, China.
Supplementary material: See accompanying pages.
J Clin Psychiatry 2016;77(5):e597-e599
dx.doi.org/10.4088/JCP.15l10156
© Copyright 2016 Physicians Postgraduate Press, Inc.
This PDF is free for all visitors!