Objective: Pharmacogenetic testing holds promise as a personalized medicine tool by permitting individualization of pharmacotherapy in accordance with genes influencing therapeutic response, side effects, and adverse events. The authors evaluated the effect on outcomes for patients diagnosed with neuropsychiatric disorders of pharmacogenetics (PGx)-guided treatment compared to usual standard of care.
Methods: This was a prospective, randomized study of 237 patients at an outpatient community-based psychiatric practice conducted between April 2015 and October 2015. Baseline patient assessments and a buccal swab were collected for pharmacogenetic testing at study initiation. For the experimental group, PGx results were provided to the clinicians as guides to treatment. Control subjects were treated according to the usual standard of care with no clinician reference to their PGx results. Neuropsychiatric Questionnaire (NPQ) and Symbol Digit Coding Test (SDC) scores and adverse drug events, hospitalizations, and medication information were collected at 30, 60, and 90 days.
Results: More than half (53%) of patients in the control group reported at least 1 adverse drug event compared to 28% of patients with PGx-guided medication management (P = .001). NPQ and SDC scores improved for both groups, but no statistical difference in efficacy as measured by these assessments was observed within the 90-day observation period.
Conclusions: Pharmacogenetic testing may facilitate psychiatric drug therapy with greater tolerability and similar efficacy compared to standard of care.
Trial Registration: ClinicalTrials.gov Identifier: NCT02411123‘ ‹’ ‹
Objective: Pharmacogenetic testing holds promise as a personalized medicine tool by permitting individualization of pharmacotherapy in accordance with genes influencing therapeutic response, side effects, and adverse events. The authors evaluated the effect on outcomes for patients diagnosed with neuropsychiatric disorders of pharmacogenetics (PGx)-guided treatment compared to usual standard of care.
Methods: This was a prospective, randomized study of 237 patients at an outpatient community-based psychiatric practice conducted between April 2015 and October 2015. Baseline patient assessments and a buccal swab were collected for pharmacogenetic testing at study initiation. For the experimental group, PGx results were provided to the clinicians as guides to treatment. Control subjects were treated according to the usual standard of care with no clinician reference to their PGx results. Neuropsychiatric Questionnaire (NPQ) and Symbol Digit Coding Test (SDC) scores and adverse drug events, hospitalizations, and medication information were collected at 30, 60, and 90 days.
Results: More than half (53%) of patients in the control group reported at least 1 adverse drug event compared to 28% of patients with PGx-guided medication management (P = .001). NPQ and SDC scores improved for both groups, but no statistical difference in efficacy as measured by these assessments was observed within the 90-day observation period.
Conclusions: Pharmacogenetic testing may facilitate psychiatric drug therapy with greater tolerability and similar efficacy compared to standard of care.
Trial Registration: ClinicalTrials.gov Identifier: NCT02411123
Prim Care Companion CNS Disord 2017;19(2):16m02036
https://doi.org/10.4088/PCC.16m02036
© Copyright 2017 Physicians Postgraduate Press, Inc.
aAltheaDx, San Diego, California
bCarolina Partners in Mental Health Care, Raleigh, North Carolina
cInovaGen, Rancho Santa Fe, California
dDepartments of Psychiatry and Community and Family Medicine, Duke University, Durham, North Carolina
*Corresponding author: Marilyn C. Olson, PhD, AltheaDx, 10578 Science Center Dr, San Diego, CA 92121 ([email protected]).
Mental disorders occur commonly in the general population and are often associated with significant adverse personal and societal costs.1 Psychiatric pharmacotherapy has traditionally relied on a trial-and-error approach, contributing to high treatment costs and poor outcomes.2 Pharmacogenetics (PGx) testing is a promising tool in personalized medicine, potentially capable of remediating the uncertainty accompanying psychiatric prescribing through analysis of genes that influence psychotropic response.
Variability in drug efficacy and toxicity can be attributed to genetic variations that impact drug-metabolizing enzymes, drug transporters, or drug targets.3-6 Significant associations also exist between genetic variants and clinical response to antidepressants,7,8 indicating that PGx testing may help inform health care providers’ medication decisions. The American Psychiatric Association9 guidelines for the treatment of major depressive disorder state that “Because of pharmacokinetic and pharmacodynamic differences among individuals, some patients may require doses higher than those approved by the FDA [US Food and Drug Administration] to achieve adequate blood levels of a medication and receive therapeutic benefit.”(p54) Additionally, recent FDA guidance on PGx testing for new drug approvals highlights the importance of PGx testing to reduce adverse events and personalize dosing.10
This naturalistic study investigated the effect of pharmacotherapy guided by the NeuroIDgenetix PGx test on therapeutic and adverse event outcomes for patients diagnosed with a range of neuropsychiatric disorders (ie, depression, anxiety, attention-deficit/hyperactivity disorder [ADHD], and psychosis) compared to that achieved with normal standard of care.
METHODS
Study Description and Participants
A total of 243 patients between the ages of 18 and 80 years were enrolled at 6 clinics throughout North Carolina affiliated with Carolina Partners in Mental Healthcare, Raleigh, North Carolina, from April 2015 to October 2015 (Clinical Trials.gov Identifier: NCT02411123). Adult patients demonstrating evidence of a neuropsychiatric disorder, including depression, anxiety, ADHD, and psychosis, requiring medication management by a qualified clinician were eligible to participate. Neuropsychiatric disorder diagnoses were assigned by the treating psychiatric clinicians on the basis of medical history and symptoms present at time of enrollment.
Prior to initiating study activities, all participants signed a consent form approved by an institutional review board (Aspire IRB). Once enrolled in the study, all subjects were monitored over a 3-month period. During the first study visit, baseline Neuropsychiatric Questionnaire (NPQ)11 and Symbol Digit Coding Test (SDC)12 scores were obtained, and trained study staff collected a buccal swab from all participating subjects and shipped the specimens to the PGx testing laboratory (AltheaDx, San Diego, California). Samples failing quality criteria for interpretation were recollected at the site. Subjects with 2 failing specimens were excluded from the study. A total of 237 of the 243 enrolled subjects had passing IDgenetix results and were randomly assigned in a 3:1 ratio of experimental to control groups. Among the 237 enrolled subjects, there were 178 subjects in the experimental group and 59 subjects in the control group.
- Inclusion of pharmacogenetic testing in medication management of patients can enhance prescribing decisions.
- Adverse drug event reduction in patients with psychiatric disorders may be achieved by implementation of pharmacogenetic testing during drug selection and dosing.
- Reduction of adverse events, as a result of pharmacogenetic testing implementation, can lead to significant savings in health care costs.
Participating clinicians were trained on the interpretation of the IDgenetix test results prior to initiating the study and were directed to consider the recommendations from the report when managing medication therapy for experimental subjects. Clinicians were trained on how to interpret the “use with caution” messages that identified genetic variants or drug-drug interactions within the report that could suggest a need for increased caution or dose adjustment.
IDgenetix test results for subjects in the control group were withheld until the end of the study, and clinicians were asked to manage control subjects as per standard of care. Patients were monitored on a monthly basis at which time the NPQ and SDC scores were reassessed, and each patient was asked about adverse drug events (ADEs) or hospitalizations since their last visit. All subjects remained blinded to their assigned study group until the end of the trial.
Pharmacogenetic Testing
Buccal swabs were collected from patients at the clinical sites and shipped to the clinical laboratory improvement amendments-certified AltheaDx laboratory (San Diego, California) within 48 hours from sample collection for PGx testing using the IDgenetix test. Medication lists reflecting patient regimen (including prescription, over-the-counter, and supplements) were submitted to the laboratory along with a test order and the collected buccal swab. This material and documentation provided the baseline information for number of medications and dose. Samples were processed, and the IDgenetix report was sent back to the clinician within approximately 72 hours after sample receipt by the AltheaDx laboratory.
Genomic DNA (gDNA) was extracted using the QIAGEN DSP DNA Midi Kit (QIAGEN, Valencia, California). The IDgenetix neuropsychiatric test panel analyzed pharmacogenetic markers across 13 genes, including CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP3A4/CYP3A5, HTR2A, HTR2C, SLC6A4, SLC6A2, COMT, ADRA2A, and MTHFR. Methods used for detection were TaqMan OpenArray Genotyping, CYP2D6 copy number variation determination, and SLC6A4 5-HTTLPR genotyping by endpoint polymerase chain reaction and capillary electrophoresis.13,14 In addition to screening for variants in these genes influential to psychotropic pharmacokinetics and pharmacodynamics, the AltheaDx proprietary IDgenetix algorithm screens for potential metabolic interactions between psychotropic treatment alternatives and medications (prescription and over-the-counter), environmental factors (food, alcohol, tobacco), and herbal products reported as taken concomitantly. Identification of such CYP450-inhibitor and CYP450-inducer effects mitigates the confounding influence of phenoconversion, phenotypic expression incongruent with genotype.
IDgenetix reports describe patient genotyping results along with a list of medications classified as “use as directed” (UAD) or “use with caution and/or increased monitoring” (UWC) on the basis of analysis findings. The medications listed in the UAD column elicited no cautions based on the analysis findings to suggest deviation from standard manufacturer-issued prescribing information.
Medications listed in the UWC column include drug-gene and drug-drug interactions identified and characterized from an extensive review of FDA-approved package inserts, professional PGx guidelines (eg, CPIC: Clinical Pharmacogenetics Implementation Consortium), and primary scientific literature. Medications in the UWC column are accompanied by brief descriptions of the reasons for caution and recommendations for appropriate clinical action. For example, the following message appears in the UWC column for citalopram for a patient with a CYP2C19 poor metabolizer phenotype: “Consider dose adjustment or alternate drug. Per CPIC guideline, consider a 50% reduction of recommended starting dose and titrate to response or select alternative drug not predominantly metabolized by CYP2C19 (CYP2C19 pm). Additionally, drug label cautions 20 mg/d maximum dose given increased QT prolongation risk.”15,16
Patient Assessments
NPQ11 and SDC12 scores were assessed at the baseline study initiation visit and at follow-up visits scheduled 30, 60, and 90 days from enrollment. The NPQ and SDC are part of a commercially available testing program called CNS Vital Signs, a computerized neurocognitive test battery for use as a routine clinical screening instrument and cognitive testing in clinical trial research.17 The NPQ is a computerized questionnaire that assesses a patient’s clinical state through a series of questions addressing symptoms of neuropsychiatric disorders.11 The short form of the NPQ includes 12 symptom domains including attention, impulsivity, memory, anxiety, panic, depression, mood stability, aggression, fatigue, sleep, suicide, and pain. Scores are reported on a scale of 0 to 300. A high NPQ score means that the patient is reporting more symptoms of greater intensity.11
The SDC is a well-established test of processing speed that is sensitive to multiple neuropsychiatric issues, including fatigue, sleep, attention, depression, and mania, and multiple neuropsychiatric disorders.12 The SDC score provides an assessment of attention and processing speed, as well as executive function. This test is sensitive to medication effects and allows monitoring of treatment.
Collection of Adverse Drug Events and Hospitalizations
At each follow-up visit, subjects were asked whether they had experienced any ADEs or had been hospitalized since their last visit. Designated study coordinators collected a description of the event as reported by the subject to the clinician; the onset, severity, and end date for the event; and the start date and name of the suspected drug all as part of a standardized ADE form. The investigator then assessed the severity of the event, how the ADE was managed, and the causality of the ADE. The possibility that the suspected drug caused the adverse event was evaluated using the following scale: unrelated/unlikely, possibly, or likely. If the reported event was consistent with those previously reported for the suspect drug as listed in the www.drugs.com database, then the ADE was classified as likely related. If the symptom was similar to a previously reported side effect, such as fatigue and unusual tiredness, then it was classified as possibly related. All others were classified as unlikely related or unrelated.
The hospitalization history questionnaire was collected at each visit and was used to record whether a subject had been hospitalized in the last 30 days. If the patient reported a hospitalization, then the length of stay, the cause for hospitalization, and whether the event started as an emergency department visit were recorded.
Economic Analysis
A post hoc economic analysis was performed to calculate the total cost of ADEs per subject using an economic model developed by Sullivan et al18 to assess the impact of ADEs on the direct costs and effectiveness of select treatments. The direct costs consisted of the drugs, office visits, and failed therapies defined as patients who discontinue treatment. The current analysis used a similar approach. On the basis of the physician rating, ADEs were classified as mild, moderate, and severe. Each ADE category and type was then “cross-walked” to overall health care cost, consisting of medical visits, and additional pharmaceutical therapies as a function of the duration of the ADE, as previously defined in the published literature.18 All costs were inflated to 2015 US dollars using the health care component of the consumer price index.
Statistical Analysis
This study was designed to assess the effectiveness of pharmacogenetic testing on the occurrence of ADEs as well as the change in NPQ and SDC scores from baseline to end of study in the experimental group compared to the control group. The endpoints of NPQ and SDC changes were assessed from baseline to 30, 60, and 90 days with the principal analysis at 90 days. The NPQ and SDC change in scores was compared between the 2 treatment arms by t test while accounting for the baseline score. The number and proportion of subjects reporting any given ADE were tabulated by treatment arm up to the 90-day visit.
RESULTS
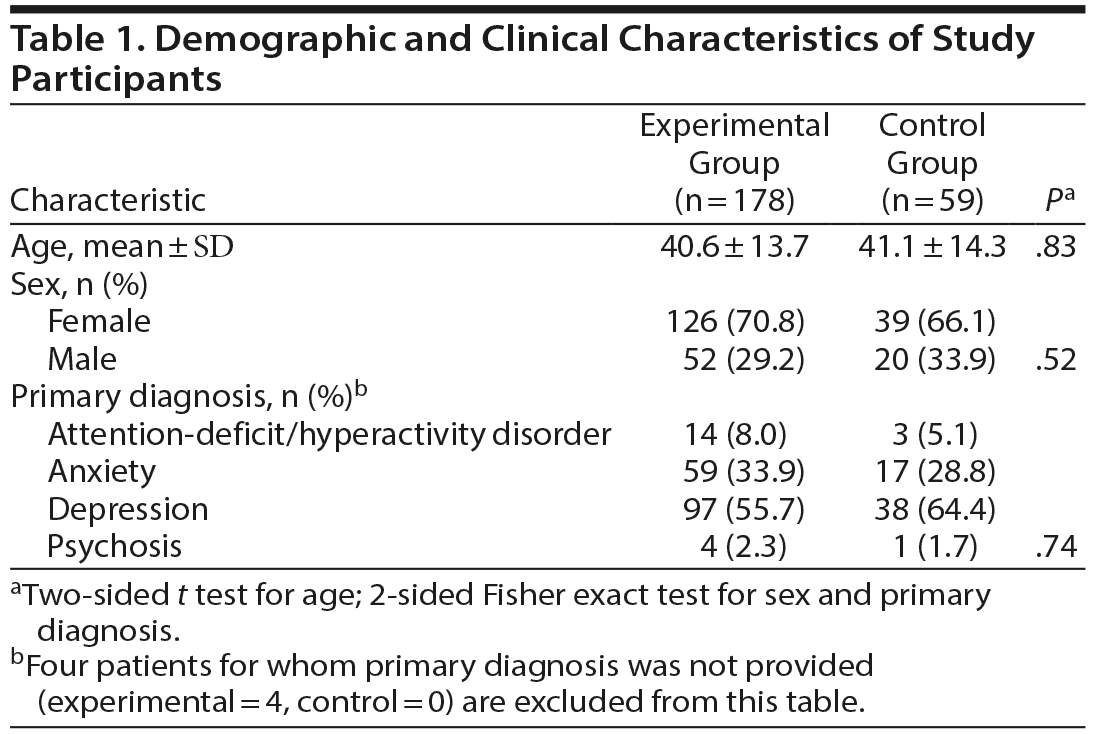
Demographic and clinical characteristics of study participants are shown in Table 1. Among the study participants enrolled and randomly assigned to the experimental group (PGx guided), the mean ± SD age was 40.6 ± 13.7 years, with 70.8% females and 29.2% males. These demographic characteristics were similar to those of the control group (P = .83). Table 1 summarizes the number of subjects with a primary diagnosis of these disorders for both the experimental and control groups. The distribution of CYP metabolizer phenotypes was similar between the experimental and control groups (Supplementary Table 1).
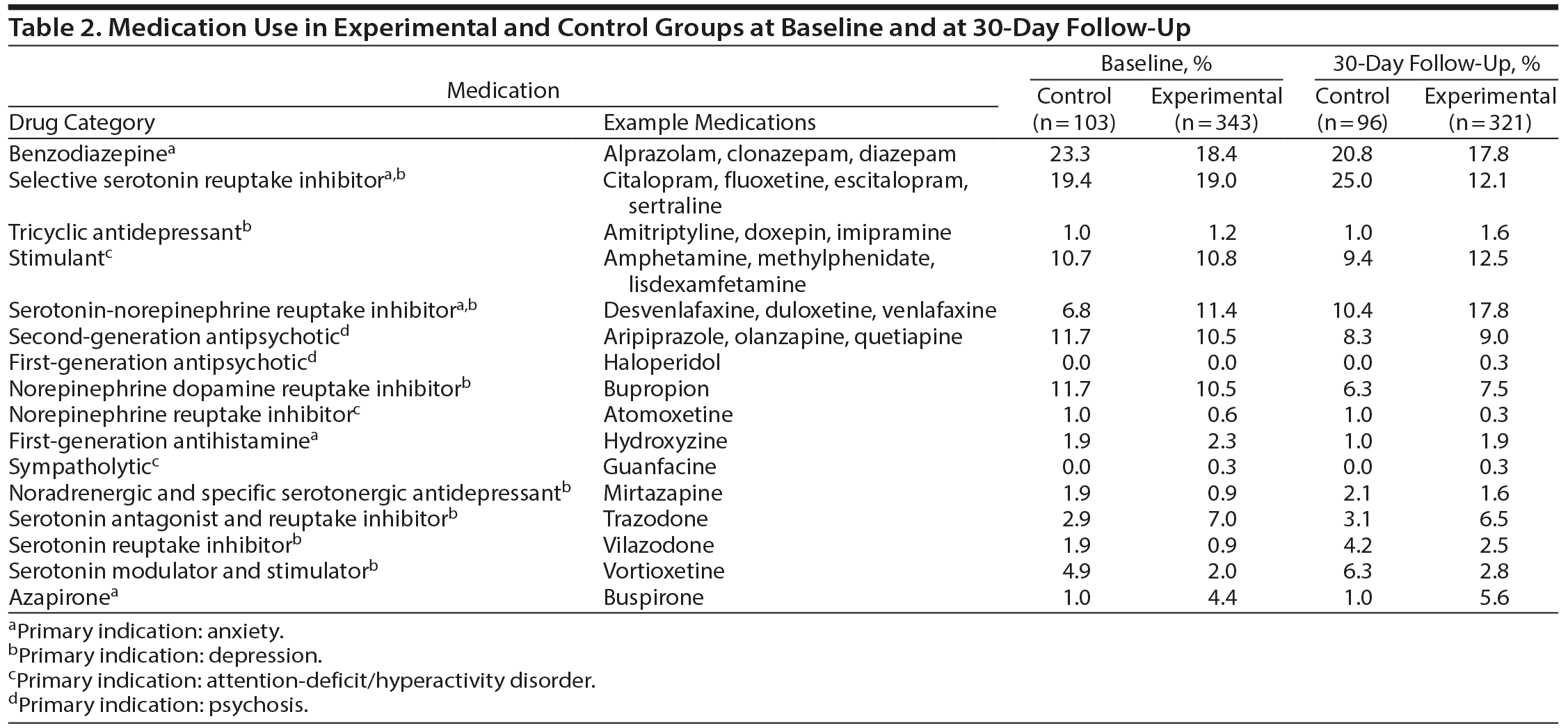
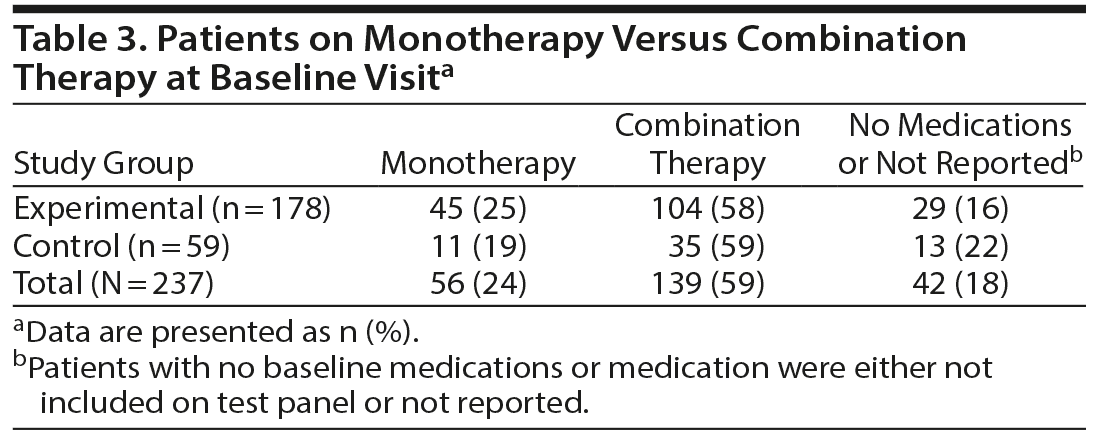
A summary of the medication use at baseline and by the 30-day follow-up visit is shown in Table 2. The total number of medications used by the study participants is indicated along with the drug category and examples of their medications. The most commonly used medications included benzodiazepines, selective serotonin reuptake inhibitors, and serotonin-norepinephrine reuptake inhibitors for treatment of depression and anxiety. The ratio of patients on monotherapy relative to those on combination therapy at baseline was similar for both the experimental and control groups (Table 3).
To assess whether clinicians were incorporating the PGx information into their management activities, an independent review of the initial regimen changes for the first 105 experimental subjects was done by a licensed PharmD (D.T.). The patients’ regimens were reviewed 30 days after enrollment with any changes evaluated in the context of their PGx reports. Results of this review revealed high clinician implementation with approximately 80% of the patient drug regimens being changed in alignment with the PGx guidance. Additionally, we tallied the percentage of UAD drugs and UWC drugs for each study group at the time of enrollment (baseline) and at 30 days to see whether implementation of our test results by the treating physicians would result in a shift toward the UAD drugs in the experimental group. At baseline, the control group exhibited a 48% UAD and 52% UWC drug use distribution, and, similarly, the experimental group had a 45% UAD and 55% UWC drug utilization ratio. At the 30-day time point, the control group drug utilization ratio was 47% UAD to 53% UWC, while the experimental group shifted to an improved ratio of 61% UAD to only 39% UWC drugs.
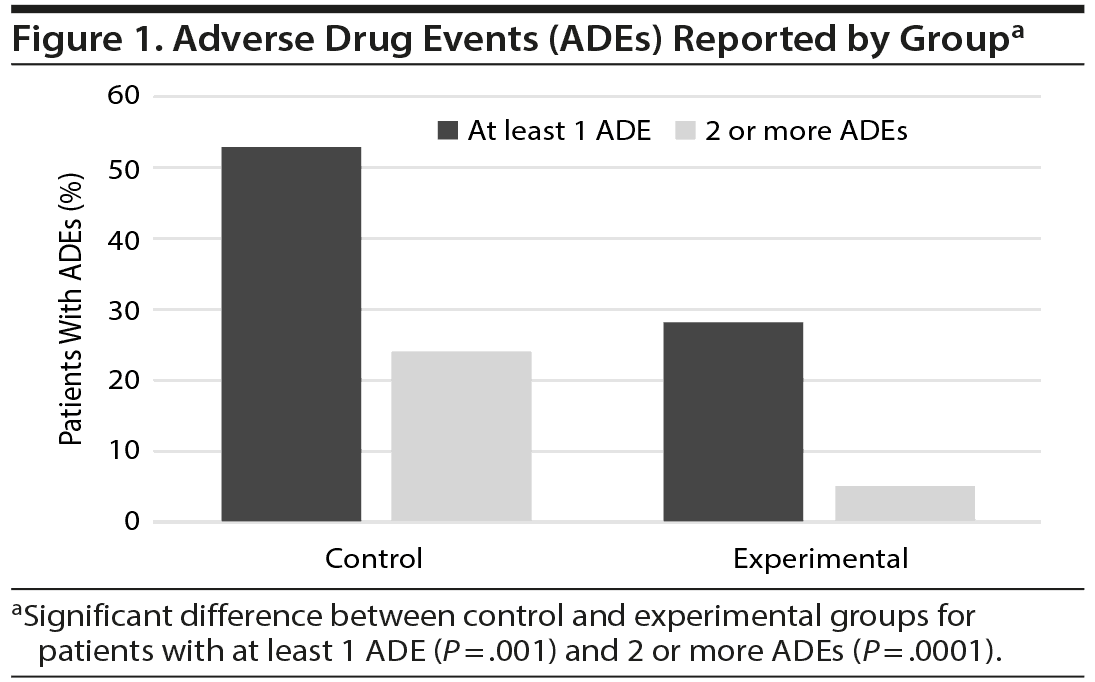
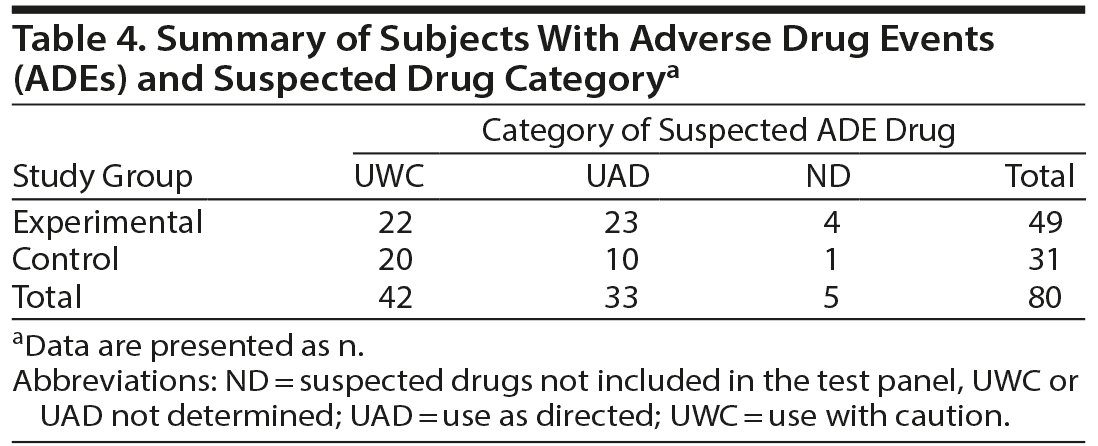
Patients were monitored for ADEs over the 3-month follow-up period. More than half (53%) of patients in the control group reported at least 1 event compared to 28% of patients with PGx-guided medication management (P = .001, Figure 1). The number of patients reporting multiple ADEs by study arm was significant, with 24% of patients in the control group reporting 2 or more ADEs during the study, compared to 5% of patients in the experimental group (P = .0001) as shown in Figure 1. Fatigue, dizziness, and nausea were the most common complaints. The impact of ADEs was evident, as it resulted in the discontinuation of the medication approximately 48% of the time overall.
There were 31 control subjects with ADEs; 20 (65%) had ADEs for which the suspected drugs were classified as UWC, and 10 (32%) had ADEs for which the suspected drugs were classified as UAD. Within the control group, twice as many subjects with ADEs were taking UWC-suspected drugs. Among the 49 experimental subjects with ADEs, 22 (45%) had ADEs for which the suspected drugs were classified as UWC, and 23 (47%) had ADEs for which the suspected drugs were classified as UAD (Table 4). It is recognized that PGx results cannot prevent all ADEs. Other patient characteristics beyond genetics such as age, comorbidities, or other factors can affect drug pharmacokinetics and pharmacodynamics contributing to ADE occurrence.
Among the medications reportedly taken by patients at the point of testing, the agents most frequently found to have drug-gene interactions were citalopram, escitalopram, fluoxetine, and sertraline. We found that there were many more medications with PGx toxicity-related warnings (53 medications for 51 subjects) compared to medications with efficacy-related warnings (14 medications for 14 subjects), which provided greater opportunities to impact ADEs in this cohort. Among the 51 subjects with toxicity-related warnings, 41 subjects had medication changes by the first follow-up time point, and 11 of the 14 subjects with efficacy-related warnings had medication changes.
While this study was not designed to quantify the costs associated with ADEs, the reduction in ADEs and related cost reductions would be expected in a similar clinical population. A post hoc economic analysis was performed to estimate the cost benefits from the reduction in ADEs. A comparison of ADE costs between the 2 study groups found that the PGx-guided group had approximately 84% lower health care costs compared to the control group (P = .019). This analysis estimated a mean annualized savings of over $1,100/patient for the PGx-guided patient group when compared to the control group.
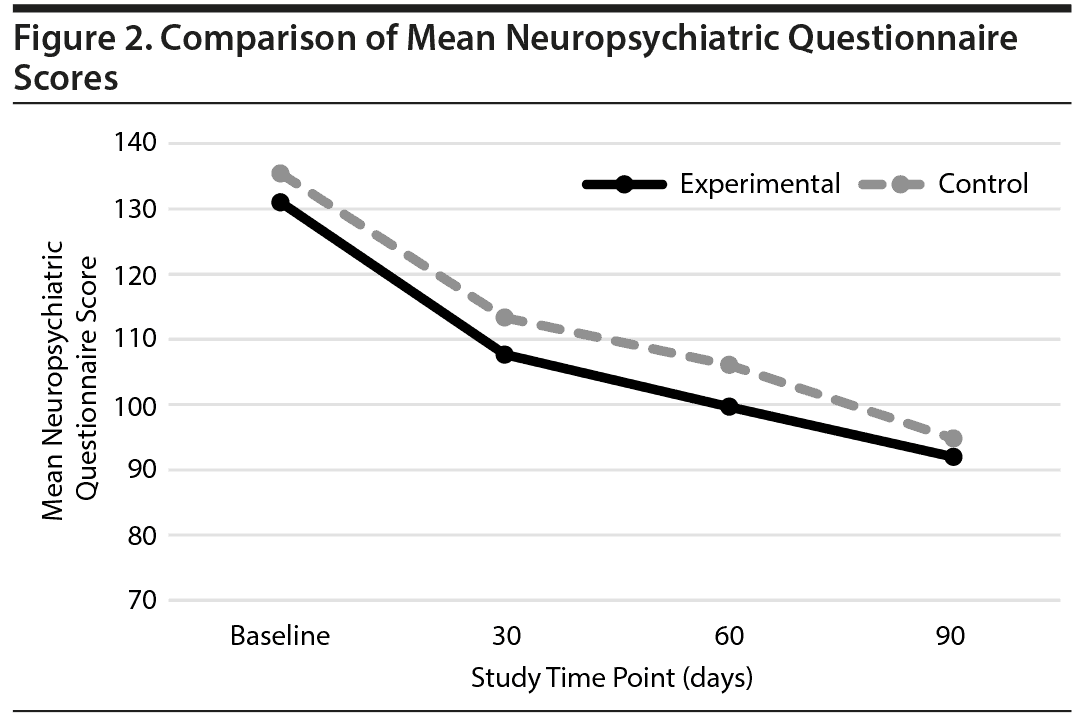
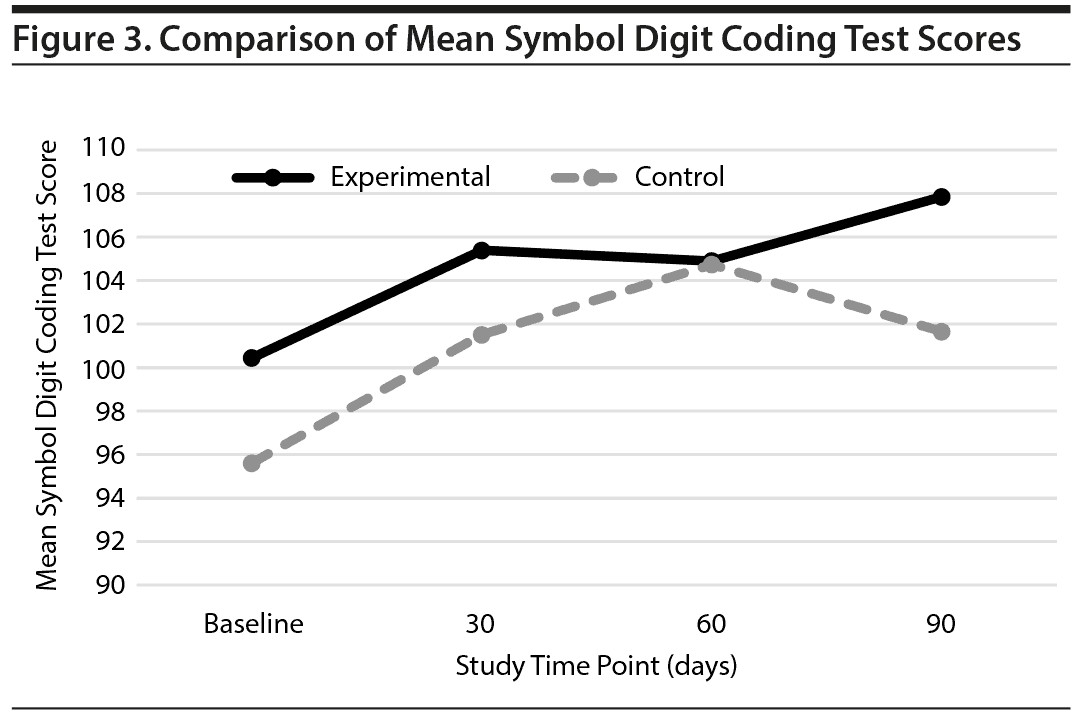
Among the 237 subjects who were enrolled in the study, 159 completed the NPQ and SDC assessments at the 3-month follow-up and were included in the SDC and NPQ analysis. Total SDC and NPQ scores were compared between the experimental and control groups. The results showed that while both groups improved over the course of the study, there was no significant difference in therapeutic efficacy between the 2 groups as assessed by these tools (Figures 2 and 3).
The overall hospitalization rates were 10% for both the experimental (18/178) and control groups (6/59). Among subjects with hospitalizations, 11 subjects reported overnight stays: 3 of the 11 overnight stays were scheduled elective procedures and 5 of the hospitalizations were recorded at baseline (occurred 30 days before enrollment). The remaining subjects were hospitalized for anxiety, depression, detoxification, bipolar disorder, and alcohol overdose. Overnight hospitalization rates were the same (5%) in the experimental and control groups.
DISCUSSION
While the body of basic research in the field of pharmacogenetics is extensive, few studies currently address its clinical utility. Initial studies,19-22 focusing on the treatment of moderate to severe depression, demonstrated a benefit in efficacy when PGx information is incorporated into the medication management of these individuals. In a retrospective study,23 investigators showed that patients who were taking a UAD medication determined by PGx had significantly lower health care visits and health care utilization costs compared to those patients who were on a UWC or “avoid use” medication. All of these studies19-22 support the notion that consideration of the individual patient’s genetic variants that are influential to drug response can help clinicians improve drug selection, resulting in improved patient outcomes and cost savings.
This study expands these findings by assessing not just major depressive disorder but other psychiatric diagnoses including anxiety disorders, ADHD, and psychotic disorders as well. We did not limit patients with depression to major depressive disorder of moderate to severe levels, and we collected data from a community psychiatric practice. We feel that these differences from previous studies allow our results to be more generalizable and representative of real-world clinical care in community settings.
To our knowledge, our study is the first to demonstrate a statistically significant improvement in ADEs with the incorporation of PGx information into the medication management of a variety of psychiatric disorders, while maintaining efficacy of treatment. Genetic variants pertinent to drug disposition can raise concerns about either efficacy, when systemic exposure to the active moiety is reduced, or tolerability, when that exposure is increased. Likewise, genetic variants affecting pharmacodynamics can impact drug efficacy or tolerability depending on their implications for a given agent’s pharmacologic activity. We found that there were many more PGx tolerability-related warnings compared to efficacy-related warnings, which provided greater opportunities to impact ADEs in this cohort. This finding most likely explains why there was a clear improvement in ADEs and no significant difference in efficacy and also aligns with the large majority of FDA drug label warnings related to tolerability versus efficacy. While this study was not designed to quantify the costs associated with ADEs, the reduction in ADEs and related cost reductions would be expected in a similar clinical population. A comparison of ADE costs between the 2 study groups found that the PGx-guided group had 84% lower health care costs compared to the control group (P = .019). fact
This study adds to the growing body of clinical utility data demonstrating the benefits of taking a more personalized approach to treating patients with psychiatric disorders. PGx test results enhanced rather than replaced traditional clinical decision-making informed by thorough patient evaluation and consideration of prior treatment history. A strength of this study is demonstration of the benefits of PGx-guided pharmacotherapy for a range of neuropsychiatric disorders. ADEs were reported for every diagnostic category except psychosis (n = 5). A weakness of the study is the relatively high dropout rate (~35%), which limited the NPQ and SDC assessments for analysis at the 3-month follow-up. Additional study weaknesses include small sample size, especially for given diagnoses, and the pragmatic design in which all variables were not controlled or well-defined.
Improvement of psychotropic regimen tolerability improves patient acceptance and adherence to treatment, thereby increasing the likelihood of therapeutic success. This study’s results demonstrate the utility of the NeuroIDgenetix test as a prescribing aid to do just that by significantly reducing ADEs.
Submitted: August 19, 2016; accepted December 23, 2016.
Published online: March 16, 2017.
Drug names: alprazolam (Xanax, Niravam, and others), aripiprazole (Abilify), atomoxetine (Strattera and others), bupropion (Wellbutrin and others), citalopram (Celexa and others), clonazepam (Klonopin and others), desvenlafaxine (Pristiq, Khedezla, and others), diazepam (Valium and others), doxepin (Silenor and others), duloxetine (Cymbalta), escitalopram (Lexapro and others), fluoxetine (Prozac and others), guanfacine (Tenex and others), imipramine (Tofranil and others), lisdexamfetamine (Vyvanse), methylphenidate (Ritalin and others), mirtazapine (Remeron and others), olanzapine (Zyprexa and others), quetiapine (Seroquel and others), sertraline (Zoloft and others), vilazodone (Viibryd).
Potential conflicts of interest: Drs Vaishnavi and Gariepy’s employer, Carolina Partners in Mental Health Care, was compensated by AltheaDx for this study. Dr Gariepy is no longer affiliated with Carolina Partners in Mental Health Care. Drs Olson, Taylor, and Garces; Mss Maciel and Cullors; and Mr Centeno are currently employed by AltheaDx. Dr Saldivar was employed by AltheaDx during the design and execution of this study.
Funding/support: This research was funded by AltheaDx.
Role of the sponsor: The sponsor (“AltheaDx” or “the company”) collaborated with Dr Vaishnavi (principal investigator [PI]) and a contracted biostatistician in the design of this study. Data collection and management was done by the PI, subinvestigators, and a clinical study coordinator (facilitated by the PI) and supported by an independent function within AltheaDx. The analysis and interpretation of data was performed by the biostatistician and the PI. Additionally, a financial consultant performed the cost analysis. Preparation, review, and approval of the manuscript was done by Dr Vaishnavi, the clinical study coordinator, and AltheaDx personnel. The PI, clinical study coordinator, biostatistician, and financial consultant are not AltheaDx customers and have no financial interest in the company. Participating personnel in this study adhered to all ethical, regulatory, and clinical research principles of good clinical practices.
Acknowledgments: The authors thank Robyn Gaylor, BA (Carolina Partners in Mental HealthCare, Raleigh, North Carolina) for her assistance with coordination of this study. Ms Gaylor has no personal affiliations or financial relationships with any commercial interest to disclose relevant to the article.
Supplementary material: See accompanying pages.
REFERENCES
1. Kessler RC, Aguilar-Gaxiola S, Alonso J, et al. The global burden of mental disorders: an update from the WHO World Mental Health (WMH) surveys. Epidemiol Psichiatr Soc. 2009;18(1):23-33. PubMed doi:10.1017/S1121189X00001421
2. Mrazek DA. Psychiatric pharmacogenomic testing in clinical practice. Dialogues Clin Neurosci. 2010;12(1):69-76. PubMed
3. Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8(4):283-289. PubMed doi:10.1097/00008571-199808000-00001
4. Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487-491. PubMed doi:10.1126/science.286.5439.487
5. Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9-39. PubMed doi:10.1146/annurev.genom.2.1.9
6. McLeod HL, Evans WE. Pharmacogenomics: unlocking the human genome for better drug therapy. Annu Rev Pharmacol Toxicol. 2001;41:101-121. PubMed doi:10.1146/annurev.pharmtox.41.1.101
7. Altar CA, Hornberger J, Shewade A, et al. Clinical validity of cytochrome P450 metabolism and serotonin gene variants in psychiatric pharmacotherapy. Int Rev Psychiatry. 2013;25(5):509-533. PubMed doi:10.3109/09540261.2013.825579
8. Porcelli S, Drago A, Fabbri C, et al. Pharmacogenetics of antidepressant response. J Psychiatry Neurosci. 2011;36(2):87-113. PubMed doi:10.1503/jpn.100059
9. Practice Guideline for the Treatment of Patients With Major Depressive Disorder, 3rd ed. American Psychiatric Association Publishing website. http://psychiatryonline.org/pb/assets/raw/sitewide/practice_guidelines/guidelines/mdd.pdf. 2010.
10. Guidance for Industry, Pharmacogenomic Data Submissions. FDA website. http://www.fda.gov/
downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm079849.pdf.
11. Gualtieri CT. An Internet-based symptom questionnaire that is reliable, valid, and available to psychiatrists, neurologists, and psychologists. MedGenMed. 2007;9(4):3. PubMed
12. Wechsler D. Wechsler Memory Scale (WMS-III). 3rd ed. San Antonio, TX: Harcourt Assessment; 1997.
13. Pratt VM, Zehnbauer B, Wilson JA, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn. 2010;12(6):835-846. PubMed doi:10.2353/jmoldx.2010.100090
14. Lesch K-P, Bengel D, Heils A, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527-1531. PubMed doi:10.1126/science.274.5292.1527
15. Hicks JK, Bishop JR, Sangkuhl K, et al; Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127-134. PubMed doi:10.1002/cpt.147
16. Celexa [package insert]. St. Louis, MO: Forest Laboratories, 2012.
17. Gualtieri CT, Johnson LG. Reliability and validity of a computerized neurocognitive test battery, CNS Vital Signs. Arch Clin Neuropsychol. 2006;21(7):623-643. PubMed doi:10.1016/j.acn.2006.05.007
18. Sullivan PW, Valuck R, Saseen J, et al. A comparison of the direct costs and cost effectiveness of serotonin reuptake inhibitors and associated adverse drug reactions. CNS Drugs. 2004;18(13):911-932. PubMed doi:10.2165/00023210-200418130-00006
19. Hall-Flavin DK, Winner JG, Allen JD, et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl Psychiatry. 2012;2:e172. PubMed doi:10.1038/tp.2012.99
20. Winner JG, Carhart JM, Altar CA, et al. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219-227. PubMed
21. Hall-Flavin DK, Winner JG, Allen JD, et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535-548. PubMed doi:10.1097/FPC.0b013e3283649b9a
22. Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin Psychopharmacol Neurosci. 2015;13(2):150-156. PubMed doi:10.9758/cpn.2015.13.2.150
23. Winner J, Allen JD, Altar CA, et al. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. PubMed doi:10.1038/tp.2013.2
This PDF is free for all visitors!